什么是LD score? 为什么需要开展LD regression analysis? 在GWAS研究中,多基因性(polygenicity,即若干较小的基因效应)和干扰因素引起的偏差(e.g.,隐性关联 cryptic relatedness,群体分层 population stratification等)都会造成检验的统计量的分布偏高(inflated) 统计量指什么?即经过卡方检验检验得到的对应SNP对目标表型的作用,用来衡量 造成association signal(SNP)对应的统计量分布偏高,是否是真的对目标表型有显著作用(有如此高的作用)? 如何将这部分不知道由于是polygenicity造成,还是由populaton stratification等不准确因素造成的effect给估计出来?答案:LD regression(实际上还有很多别的模型来做这这个heritability估计) 通过LD score regression,我们可以通过研究检验统计量与连锁不平衡(linkage disequilibrium)之间的关系来定量分析每部分的影响,即对干扰因素平均效应的估计值进行计算 LD Score的计算原理是什么? GWAS检验中,对一个SNP效应量的估计通常也会包含与该SNP成LD的其他SNP的效应,也就是说一个与其他SNP呈现高LD的SNP,通常也会有更高的卡方检验量。 LD regression的原理是什么? 输入文件:要求PLINK处理过后的genotype数据 作者推荐的LD Score估计遗传范围(选择用于计算LD Score遗传距离的窗口):1 centiMorgan (cM) window 可以使用 在针对MHC等复杂区域(e.g.,LD延展度更高的region),需要设置更大的window size;具有比较高的recombination rate就不需要设置太大的window size It is sensible to use a larger window (as measured in kb) in regions like the MHC where LD spans over tens of megabases than in regions with high recombination rate, where LD doesn't extend beyond ~100kb. 以LDSC官方提供的 包含所使用的cml以及基本运行结果, 第一个文件包含了目标region所包含的SNP数目 第二个文件包含了过滤MAF(>5%)的SNP数目 LDSC一般基于后者来进行heritability的估计 需要的输入文件, 结果文件解读包含:heritability、genetic correlation、LD regression估计出来的intercetp(衡量了polygenicity之外的因素对SNP effect的影响)等 转换之后生成 log文件的最后一部分给出了summary statistics的meta信息, 举个例子, 与使用 作者对此的解释:总的来说就是所使用的SNP dataset不同,rg由于需要同时计算genetic correlation,所以并不是直接使用对应表型的SNP dataset 以 Lambda GC,代表了估计结果受到population stratification的影响程度 计算公式:, 0.4549,the expected median of a variable from the distribution ,通过比较case和control中的genotype得到的卡方统计量 Ratio(attenuation ratio):,用于衡量在LD regression中,polygenicity之外的因素对(理想情况下,应为0)SNP effect的影响() genetic covariance: genetic correlaton: 由于ldsc估计出来的heritability是obseved scale,为了研究之间的可比较性,需要转换到liability scale,写在前面
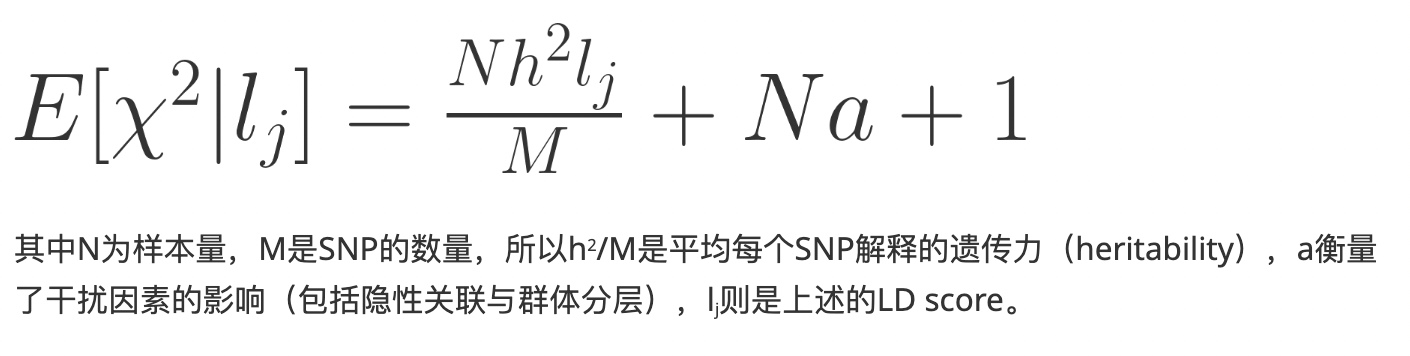
LDSC|安装
git clone https://github.com/bulik/ldsc.git
cd ldsc
mamba env create --file environment.yml
mamba activate ldsc
LDSC|LD Score Calculation
.bed/.bim/.fam作为输入文件,可以使用PLINK VCF来生成plink --cm-map来保留遗传距离1kg_eur.tar.bz2来说明toy data的结果。01)Univariate LD Score
python ldsc.py\
--bfile\
--l2\
--ld-wind-cm 1\
--out 22
--l2,指示计算LD Score--ld-wind-cm代表选择的window size(基于遗传距离),还可以直接基于物理距离、SNP个数等结果文件解读
1)22.log
Beginning analysis at Fri Jan 30 10:58:44 2015
Read list of 19156 SNPs from 22.bim
Read list of 379 individuals from 22.fam
Reading genotypes from 22.bed
After filtering, 19156 SNPs remain
~~~Estimating LD Score.~~~
Writing LD Scores for 19156 SNPs to 22.l2.ldscore.gz
2)
22.l2.M, 22.l2.M_5_50
3)22.l2.ldscore.gz
gunzip -c 22.l2.ldscore.gz | head
CHR SNP BP L2
22 rs9617528 16061016 1.271
22 rs4911642 16504399 1.805
22 rs140378 16877135 3.849
22 rs131560 16877230 3.769
22 rs7287144 16886873 7.226
22 rs5748616 16888900 7.379
22 rs5748662 16892858 7.195
22 rs5994034 16894090 2.898
22 rs4010554 16894264 6.975
02)Partitioned LD Score
LDSC|LD regression
01)genetic correlation & heritability
1)数据下载 & 格式转换
#
wget www.med.unc.edu/pgc/files/resultfiles/pgc.cross.bip.zip
wget www.med.unc.edu/pgc/files/resultfiles/pgc.cross.scz.zip
wget https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2
wget https://data.broadinstitute.org/alkesgroup/LDSCORE/w_hm3.snplist.bz2
# 由于GWAS summary statistics不能直接用于ldsc的计算,需要转换
munge_sumstats.py \
--sumstats pgc.cross.SCZ17.2013-05.txt \
--N 17115 \
--out scz \
--merge-alleles w_hm3.snplist
munge_sumstats.py \
--sumstats pgc.cross.BIP11.2013-05.txt \
--N 11810 \
--out bip \
--merge-alleles w_hm3.snplist
.log文件,包含的信息:使用的命令以及header对应的信息以及对summary statistics过滤的condition信息
INFO > 0.9, MAF > 0.01 and 0 < P <= 1.,去除了INDELs、strand ambiguous SNPs、SNPs with duplicated rs numbersMetadata:
Mean chi^2 = 1.229
Lambda GC = 1.201
Max chi^2 = 32.4
11 Genome-wide significant SNPs (some may have been removed by filtering).
Conversion finished at Mon Apr 4 13:21:29 2016
Total time elapsed: 16.07s
Reading list of SNPs for allele merge from w_hm3.snplist
Read 1217311 SNPs for allele merge.
Reading sumstats from pgc.cross.SCZ17.2013-05.txt into memory 5000000.0 SNPs at a time.
Read 1237958 SNPs from --sumstats file.
Removed 137131 SNPs not in --merge-alleles.
Removed 0 SNPs with missing values.
Removed 256286 SNPs with INFO <= 0.9.
Removed 0 SNPs with MAF <= 0.01.
Removed 0 SNPs with out-of-bounds p-values.
Removed 2 variants that were not SNPs or were strand-ambiguous.
844539 SNPs remain.
Removed 0 SNPs with duplicated rs numbers (844539 SNPs remain).
Using N = 17115.0
Median value of or was 1.0, which seems sensible.
Removed 39 SNPs whose alleles did not match --merge-alleles (844500 SNPs remain).
Writing summary statistics for 1217311 SNPs (844500 with nonmissing beta) to scz.sumstats.gz.
2)遗传力估计 + 遗传相关性估计
# rg
ldsc.py \
--rg scz.sumstats.gz,bip.sumstats.gz \
--ref-ld-chr eur_w_ld_chr/ \
--w-ld-chr eur_w_ld_chr/ \
--out scz_bip
# h2
ldsc.py \
--h2 scz.sumstats.gz \
--ref-ld-chr eur_w_ld_chr/ \
--w-ld-chr eur_w_ld_chr/ \
--out scz_h2
参数解读
--rg,计算genetic correlation(作者给出的例子中直接以SCZ和bipolar为例)
--h2来计算得到的heritability存在不同
--w-ld|--w-ld-chr,在进行的LD回归的估算时,理论上要考虑到该SNP和别的SNP的之和,但是在实际中LD回归对SNP的权重不敏感,所以使用--ref-ld-chr即可结果解读
--rg的分析结果为例
Intercept,the LD Score regression interceptHeritability of phenotype 1
---------------------------
Total Observed scale h2: 0.5907 (0.0484)
Lambda GC: 1.2038
Mean Chi^2: 1.2336
Intercept: 1.0014 (0.0113)
Ratio: 0.0059 (0.0482)
Heritability of phenotype 2/2
-----------------------------
Total Observed scale h2: 0.5221 (0.0531)
Lambda GC: 1.1364
Mean Chi^2: 1.1436
Intercept: 1.0013 (0.0094)
Ratio: 0.0093 (0.0652)
Genetic Covariance
------------------
Total Observed scale gencov: 0.3644 (0.0368)
Mean z1*z2: 0.1226
Intercept: 0.0037 (0.0071)
Genetic Correlation
-------------------
Genetic Correlation: 0.6561 (0.0605)
Z-score: 10.8503
P: 1.9880e-27
02)将heritability结果转换到liability scale
ldsc.py \
--rg scz.sumstats.gz,bip.sumstats.gz \
--ref-ld-chr eur_w_ld_chr/ \
--w-ld-chr eur_w_ld_chr/ \
--out scz_bip \
--samp-prev 0.5,0.5 \
--pop-prev 0.01,0.01
参考资料
资源
Hi,这里是有朴的第二大脑。
很高兴与你相遇
很高兴与你相遇